Applied Biosystems 4000 Series (TOF/TOF)
The Applied Biosystems 4000 Series instruments are unusual in that the spectra are
stored in an Oracle database, rather than separate files for each experiment. Applied
Biosystems provide two software packages to support the 4700 and 4800 TOF/TOF
instruments; 4000 Series Explorer and GPS Explorer. In addition, individual spectra
can be exported as *.T2D files and opened by Data
Explorer.
GPS Explorer provides extensive Mascot integration. Search parameters are defined as part of
an Analysis in GPS Sample Setup. On completion of each search, the results are imported
into the 4000 series database and can be viewed and reported using the GPS Results Browser.
The workflow supports both ICAT® and iTRAQTM quantitation.
Interactive Searching
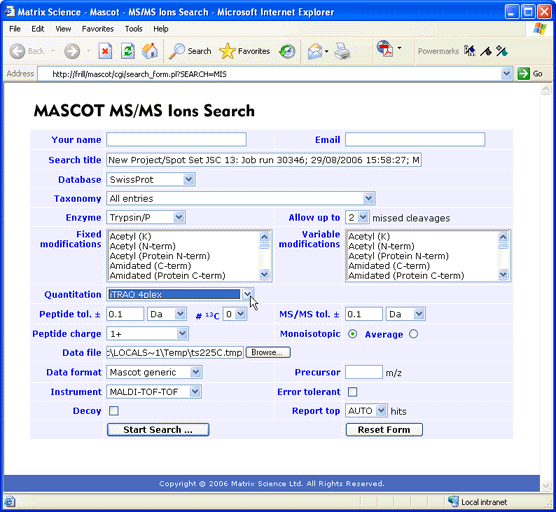
If you wish to submit a search from a web browser using the Mascot search form, the 4000
Series Explorer has a function called Peaks to Mascot that exports a peak list to an MGF
file.
Another way to submit a search through the standard search form is to use a free utility
called TS2Mascot. This will run under Microsoft Windows NT 4.0 SP6, Windows 2000, Windows XP Professional,
and Windows 2003 Server (32 bit only). TS2Mascot communicates with the Applied Biosystems 4000
Series database using ODBC. The PC will need to have Oracle client software installed,
including the Oracle ODBC driver. To submit a Mascot search from TS2Mascot, Microsoft
Internet Explorer 4.01 SP2 or later must be installed and operational.
To install TS2Mascot, run the
Windows installer package.
Unless 4000 Series Explorer or GPS Explorer is already installed
on the target PC, you may need to create a local Oracle service name,
as described in the
setup instructions.
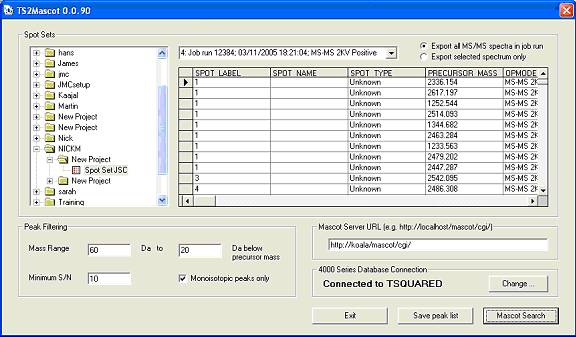
TS2Mascot is very simple to use. It is launched from the Windows Start menu, (Programs; Mascot; TS2Mascot).
The folder tree in the Spot Sets frame is populated with project folders from the 4000 series database.
If you have more than one 4000 series database, choose Change to connect to a different database.
Navigate the folder tree and select a spot set icon. This will populate the adjacent drop down list
with the job runs for the spot set. The grid area displays details of individual job run items
for the selected job run.
Peak filtering settings are equivalent to those used in Applied Biosystems GPS Explorer.
The mass range limits only apply to MS/MS fragment ion peaks.
The contents of the Peak Filtering and Mascot Server URL fields are sticky. If you change them,
then process a spot set, the new values will be remembered.
Save peak list invokes a standard file selection dialog before processing the data.
Before choosing Mascot Search, ensure that you have entered a valid URL for a Mascot
server. You can enter the URL of the Matrix Science public web site, but remember that
this has a limit of 300 spectra in a single search.
While the peak list is being created, a progress bar is displayed, and all the
controls are disabled apart from a Cancel button. Once peak list export is complete,
if you have chosen Mascot Search, the default web browser will be launched and the
Mascot MS/MS search form displayed. The search title and data file path are filled in
automatically. The other settings are the search form defaults, (which can be customised
by following a link on the search form selection page).
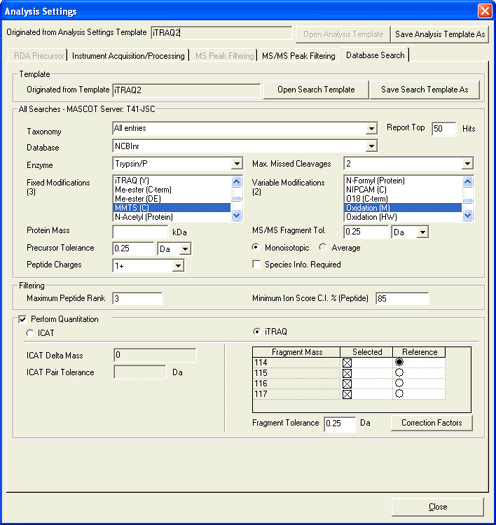
If you want to perform iTRAQ quantitation outside of GPS Explorer, it is important to use TS2Mascot
because the peak areas in the peak list exported by Peaks to Mascot or submitted to a Mascot server by GPS
Explorer are not quite the same as those required for quantitation. For a detailed description of Mascot
support for iTRAQ quantitation, refer to the quantitation help page for the
Reporter Protocol.
Automation using Mascot Daemon
Mascot Daemon can use TS2Mascot as a data import filter. First create a suitable parameter
set and save it. If the parameter set is for searching MS/MS data, the file format should be specified as
Mascot Generic.
In the Task editor, if you want to use TS2Mascot as the data import filter, select the parameter
set and then choose the AB 4000 Series TS2Mascot option from the data import filters.
The Add Files ... button invokes a database browser window.
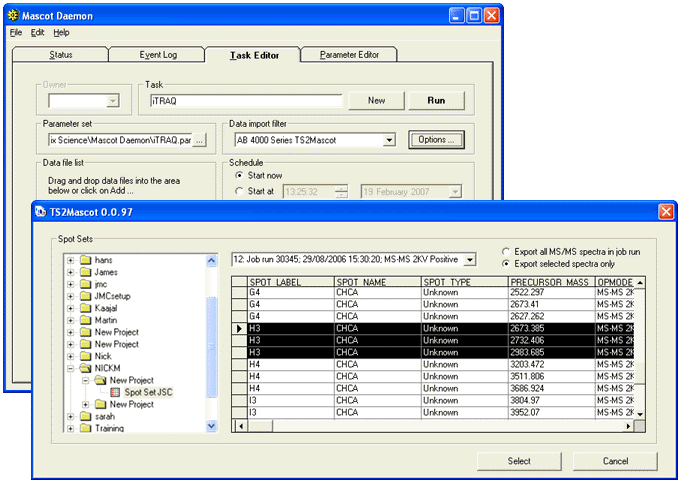
Choose Export all MS/MS spectra in job run for a single search of all the MS/MS spectra in the job run.
Any row selection in the data grid has no effect when this option is used.
Alternatively, choose Export selected spectra only and select one or more rows
in the data grid to search these spectra as separate searches, (e.g. a batch of PMFs).
This option works for both MS and MS/MS data.
|