Scoring Schemes
The Mowse scoring algorithm is described in [Pappin, 1993].
The first stage of a Mowse search is to compare the
calculated peptide masses for
each entry in the sequence database with the set
of experimental data. Each calculated value which falls within
a given mass tolerance of an experimental value counts as a match.
A molecular weight range for the intact protein can be used as
a pre-filter.
Rather than just counting the number of matching peptides,
Mowse uses empirically determined factors to assign a statistical
weight to each individual peptide match. The matrix of weighting
factors is calculated during the database build stage, as follows:
A frequency factor matrix, F, is created, in which each
row represents an interval of 100 Da in peptide mass, and each
column an interval of 10 kDa in intact protein mass. As each sequence
entry is processed, the appropriate matrix elements fi,j
are incremented so as to accumulate statistics on the size distribution
of peptide masses as a function of protein mass. The elements
of F are then normalised by dividing the elements of each
10 kDa column by the largest value in that column to give the
Mowse factor matrix M:
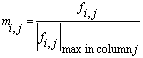
After searching the experimental mass values against a calculated
peptide mass database, the score for each entry is calculated
according to:
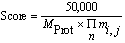
Where MProt is the molecular weight of the
entry and the product term is calculated from the Mowse factor
elements for each match between the experimental data and peptide
masses calculated from the entry.
Mascot incorporates a probability based implementation of the Mowse
algorithm. The Mowse algorithm is an excellent starting point because it
accurately models the behaviour of a proteolytic enzyme. By casting the
Mowse score into a probabilistic framework, we gain a number of additional
benefits:
- A simple rule can be used to judge whether a result is significant
or not.
- Different types of matching (peptide masses and fragment ions)
can be combined in a single search.
- Scores from different searches and on different databases can be compared.
- Search parameters can be optimised more readily by iteration.
Matches using mass values (either peptide masses or MS/MS fragment ion masses)
are always handled on a probabilistic
basis. The total score is the absolute probability that the observed match is a
random event. Reporting probabilities directly can be confusing. Partly because they
encompass a very wide range of magnitudes, and also because a "high" score is a
"low" probability, which can be ambiguous. For this reason, we report scores as -10*LOG10(P), where P is the absolute
probability. A probability of 10-20 thus becomes a score of 200.
Significance Level
Given an absolute probability that a match is random, and knowing the size
of the sequence database being searched, it becomes possible to provide
an objective measure of the significance of a result. A commonly accepted
threshold is that an event is significant if it would be expected to occur
at random with a frequency of less than 5%. This is the value which is reported
on the master results page.
The master results page for typical peptide mass fingerprint search
(open in new window)
reports that "Scores greater than 67 are significant
(p<0.05)". The histogram of the score distribution looks like this:
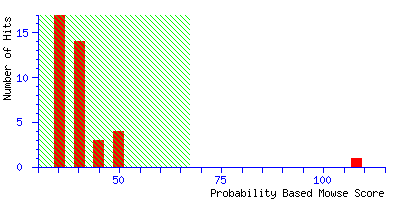
The protein with the high score of 108 is a 26 kDa heat shock protein from yeast.
This is a nice result because the highest score is highly significant, leaving little
room for doubt.
(It may be useful to think of the score histogram as a highly magnified view of the extreme
tail of the distribution of scores for all the entries in the sequence database. In this case,
50 entries out of 257,964. Scores in the green region are inside this tail, and are of
no significance. A real match, which is a non-random event, gives a score which is well clear
of the tail.)
It is important to distinguish between a significant
match and the best match. Ideally, the correct match is both the best match
and a significant match. However, significance is a function of
data quality. It may be that there are just not enough mass values or the
mass measurement accuracy is not good enough to get a significant
match. This doesn't mean that the best match isn't correct, it just means
that you must study the result more critically.
To illustrate the difference between a significant match and a correct match,
try repeating the search in the example, but with the mass tolerance
increased from ±0.1 Da to
±1.0 Da. The discrimination of the search is greatly reduced, and the score
for the correct match falls close to the significance level:
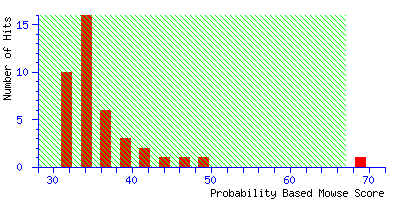
The best match is still correct, but it is barely significant. If we did 20 such searches,
we could expect to get this score by chance alone because there is such a huge number of entries
in the sequence database. The correct match remains at the top of the hit list even when the
mass tolerance is increased to ±2.0 Da, but because the score is well below the significance
threshold, there could be no confidence in this match if it was an unknown. Increase the mass
tolerance to ±2.5 Da, and the match is finally lost. The highest score is 48 for a random
match
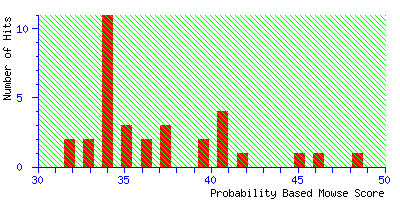
Even if this was an unknown, it is clear from the significance level that this is not a useful
match, and there is no danger of this result becoming a false positive.
Expectation Values
Each protein score in a peptide mass fingerprint, and each ions score in an MS/MS search,
is accompanied by an expectation value. This is the number of matches with
equal or better scores that are expected to occur by chance alone.
It is directly equivalent to the
E-value
in a Blast search result.
For a score that is exactly on the default significance threshold, (p<0.05), the expectation
value is also 0.05. Increase the score by 10 and the expectation value drops to 0.005.
The lower the expectation value, the more significant the score.
If the number of matched mass values is constant, the score in a peptide mass
fingerprint will be inversely related to the
mass tolerance, as shown in the example above. This is not the case for an
MS/MS ions search, where increasing the peptide mass tolerance will have no
effect on the ions score. This is
because the ions score comes from the MS/MS fragment ion matches.
Opening up the peptide mass tolerance means that Mascot has to test many more
peptides, so the search takes longer and the discrimination is reduced, but the
ions score remains unchanged.
Of course, if the peptide mass tolerance is set too tightly,
in an effort to improve discrimination, one or more of the peptide matches
may be lost, which will dramatically reduce the overall score.
Like any statistical approach, Probability Based scoring depends
on assumptions and models.
One of these assumptions is that the entries in the
sequence databases are random sequences. This is not always a good
assumption. Some of the most glaring examples involve extended repeats, such as
AAC62527,
porcine submaxillary apomucin. Although the molecular weight of
this protein is 1.2 MDa, over 80% of the sequence is composed of an identical 7 kDa
repeat. It is difficult to know how to treat such cases. If a single experimental
peptide mass is allowed to match to multiple calculated masses, then a single
experimental mass which matches within a repeat will give a huge and
meaningless score. But, if duplicate matches are not permitted, it
will be virtually impossible to get a match to such a protein because the number
of measurable mass values is too small to give a statistically significant score.
Another assumption is that the experimental measurements are independent
determinations. This will not be true if the data include
multiple mass values for the same peptide, even if
these are from ions with different charge states in an electrospray LC-MS run. Good
peak detection and thresholding (in both mass and time domains for LC-MS) are
essential for any scoring algorithm to give meaningful results.
Amino acid sequence or composition information, if included as
seq(…) or comp(…)
qualifiers, is treated as a filter on the candidate sequences.
Ambiguous sequence or composition data can be used (in a manner similar
to a regular expression search in computing) but it still functions as a filter, not
a probabilistic match of the type found in a Blast or Fasta search.
In contrast, tag(…) and
etag(…) qualifiers are scored probabilistically.
That is, the more qualifiers that match, the higher the score, but all qualifiers are not
required to match.
|