Sequence Database Setup: IPI
Overview
IPI (International Protein Index) is compiled by the
EBI (European Bioinformatics Institute)
to provide a top level guide to the main databases that describe the human and mouse proteomes:
SWISS-PROT,
TrEMBL,
NCBI RefSeq and
Ensembl. The aim is to:
- effectively maintain a database of cross references between the primary data sources
- provide a minimally redundant yet maximally complete set of proteins (one sequence per transcript)
- maintain stable identifiers (with incremental versioning) to allow the tracking of sequences in IPI between IPI releases.
IPI is updated monthly in accordance with the latest data released by the primary data sources.
There are currently two IPI databases, Human and Mouse. This document uses the Human database as an example.
To work with the Mouse database, simply substitute the word "mouse" for "human". For example,
the compressed Fasta file is ipi.MOUSE.fasta.gz, the db_update.pl keyword is IPI_mouse_from_EBI, the recommended
Mascot name is IPI_mouse, etc.
Download
ftp://ftp.ebi.ac.uk/pub/databases/IPI/current/
for the latest release.
ftp://ftp.ebi.ac.uk/pub/databases/IPI/old/
for earlier releases.
There are two files: a Fasta database file (ipi.HUMAN.fasta.gz) and a reference file in Swiss-Prot format
(ipi.HUMAN.dat.gz).
It is worth getting the reference file because then you can view a full text report, including cross
reference information, without linking out to the internet.
To download updates automatically, the relevant definition block in
db_update.pl is IPI_human_from_EBI:
Taxonomy
Taxonomy is not required because all entries are from the same species
Parse Rules
A typical Fasta title line is:
>IPI:IPI00177321.1|REFSEQ_XP:XP_168060
Tax_Id=9606 similar to NOD3 protein
The IPI accession number is the preferred identifier. In most cases, it is not necessary
to include the version number.
Accession from Fasta title: ">IPI:\([^| .]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
The corresponding line in the Dat file is:
ID IPI00177321.1 IPI; PRT; 316 AA.
Accession from Ref file: "^ID \([^ .]*\)"
Configuration
For this example, both database files were downloaded to
C:\Inetpub\MASCOT\sequence\IPI_human\current,
decompressed using gzip,
and renamed to IPI_human_2.31.dat and IPI_human_2.31.fasta.
When updating an active database, it is important to rename the Fasta file last, because Mascot
will begin database exchange as soon as it sees a new Fasta file that matches the wildcard path for
the database.
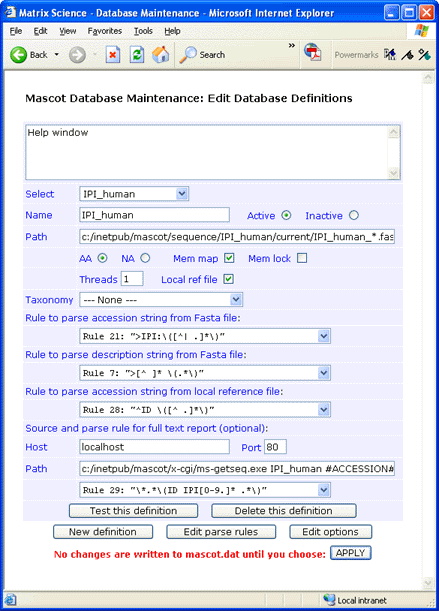
If you prefer not to have the reference file locally, full text for individual entries can be retrieved across the web
from the EBI SRS server. For an SRS6
server, the syntax for the Path field is:
/srs6bin/cgi-bin/wgetz?-e+[IPI-acc:#ACCESSION#]+-vn+2
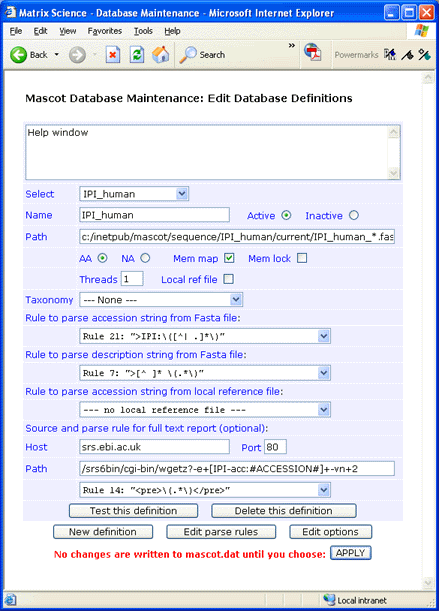
Make sure that the final parse rule has the correct case. Early versions of wgetz return HTML pages tagged
with <PRE>, while later versions use <pre>. Parse rules are always case sensitive.
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|