Sequence Database Setup: TrEMBL
| Trembl Release 34.0 |
| The syntax of the Fasta title line changed in Trembl
release 34.0. This page has been updated to illustrate the new parse rules.
|
|
Overview
TrEMBL is a computer-annotated supplement
of SWISS-PROT that contains all the translations of EMBL nucleotide sequence entries not yet integrated in
SWISS-PROT.
TrEMBL is developed by the SWISS-PROT groups at
SIB and
EBI.
Download
Expasy:
ftp://ftp.expasy.org/databases/uniprot/knowledgebase
EBI:
ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase
The EBI site mirrors the Expasy site. The relevant files are:
- Version info: reldate.txt
- TrEMBL Fasta file: uniprot_trembl.fasta.gz
- TrEMBL Dat file: uniprot_trembl.dat.gz
To download TrEMBL updates automatically, the relevant definition block in
db_update.pl is Trembl_complete_from_EBI.
Taxonomy
Taxonomy for the TrEMBL Dat file is identical to that for SWISS-PROT, and is predefined
in mascot.dat as "Swiss-prot DAT". The following taxonomy files are required:
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase/docs/speclist.txt
Note that the taxonomy files go into the taxonomy directory, not into the sequence database
directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
Parse Rules
A typical Trembl Fasta title line is:
>O05473|O05473_SULIS ORF67 - Sulfolobus islandicus
You can use either the ID (O05473_SULIS) or
the AC (O05473) as the identifier.
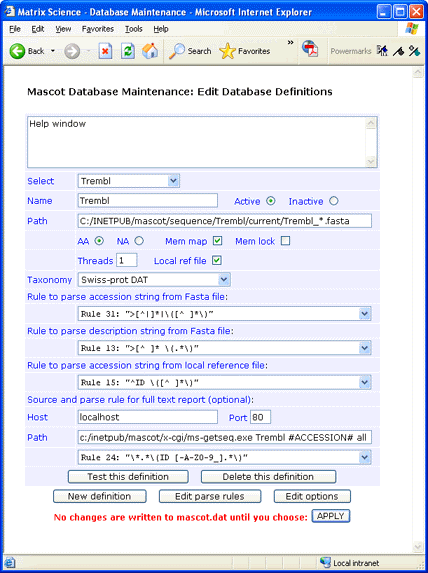
ID from Fasta title: ">[^|]*|\([^ ]*\)"
AC from Fasta title: ">\([^|]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
The corresponding line in the Dat file is:
ID O05473_SULIS Unreviewed; 67 AA.
ID from Ref file: "^ID \([^ ]*\)"
Configuration
For this example, the database files were downloaded to
C:\Inetpub\MASCOT\sequence\Trembl\current,
decompressed using gzip,
and renamed to Trembl_34.1.dat and Trembl_34.1.fasta.
When updating an active database, it is important to rename the Fasta file last, because Mascot
will begin database exchange as soon as it sees a new Fasta file that matches the wildcard path for
the database.
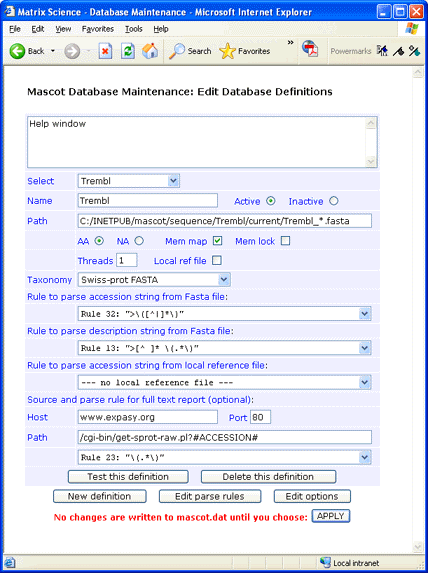
If you decide not to have the reference file locally, full text for individual entries can be retrieved across the web
from an SRS server or Expasy. This can be done using either the ID or the AC as the identifier.
For Expasy, the syntax for the Path field is:
/cgi-bin/get-sprot-raw.pl?#ACCESSION#
Where #ACCESSION# represents either the AC or ID. For an SRS
server, the syntax for the Path field is:
Retrieve by ID: /srsbin/cgi-bin/wgetz?-e+[UNIPROT-id:#ACCESSION#]+-vn+2
Retrieve by AC: /srsbin/cgi-bin/wgetz?-e+[UNIPROT-acc:#ACCESSION#]+-vn+2
This screen shot illustrates a configuration in which the identifier is AC and there is no local Dat file:
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|