Sequence Database Setup: Database Maintenance
The Database Maintenance utility provides an interface to the main configuration file, mascot.dat.
Note that no changes are made to mascot.dat until you explicitly choose to 'Apply' the new definitions.
Whenever changes are applied, the Database Maintenance script will automatically make backup copies
of mascot.dat. These are numbered sequentially from mascot.dat.1 onwards. If you have problems after
making changes, just revert to the most recent backup.
You can also edit mascot.dat using any text editor, but first familiarise yourself with Chapter 6
in the Setup and Administration manual, Configuration and Log Files, which contains a comprehensive
description of the configuration parameters.
Databases definitions form
This form can be used to modify existing database definitions or add new ones. When Mascot is first installed,
the only active database is MSDB, but several other databases are pre-defined. Individual database configurations
can be displayed by selecting them from the drop-down list near the top of the form.
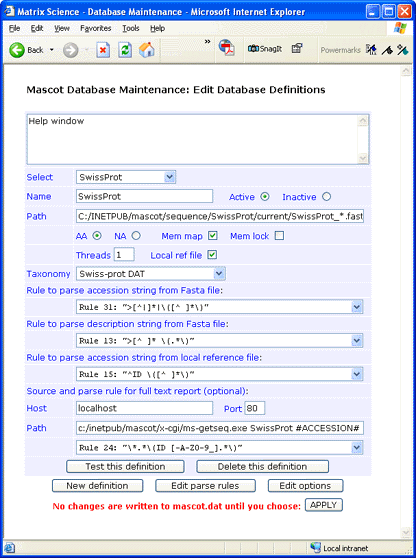
Name: Each database must have a unique name. Ideally, the name should be short and descriptive.
Note that these names are case sensitive, and much confusion can be caused by creating (say) Sprot and SPROT.
Active / Inactive: A database that is no longer required can be marked as inactive so as to retain
the definition in mascot.dat. If you are certain that you will never need the definition again, it can be
deleted using the button at the bottom of the definition block.
Path: The location of the FASTA file is defined in the Path field. This must be the fully qualified
path to the FASTA file, with a wild card in the filename, (not the extension). The delimiters between directories
must always be forward slashes, even if Mascot is running on a Windows system. The filename does not have to be
based on the database name, but doing so can make administration easier.
AA / NA: A pair of radio buttons is used to specify whether the database is amino acid (protein) or
nucleic acid.
Mem map: Database files should always be memory mapped. Unlike memory locking, this does not consume
physical RAM.
Mem lock: Memory mapped files can be locked in memory, but only if the computer has sufficient RAM.
Having a database locked in memory means that it can never be swapped out to disk, ensuring maximum possible
search speed. If you try to lock databases into RAM when there isn't room, this will not be a major problem.
The locking will fail, generate an error message, and Mascot will carry on regardless. A more serious problem
is when there is just sufficient RAM to lock the databases, but none left over for searches or other applications.
In this case, the whole system will slow down and the hard disk will be observed to be "thrashing". Eventually,
the system is likely to hang or crash.
Threads: A Mascot search can use multiple threads. If you are running in cluster mode, 'Threads'
must be set to 1. Otherwise, specify the same number of threads as the number of processors in your license.
Local ref file: The FASTA database file must be available locally. For certain databases
(e.g. MSDB, IPI, Trembl, SwissProt), it is also possible to have a local reference file, from which full text
information can be taken for a 'Protein View' report. If you have a local copy of the reference file,
check the 'Local ref file' box. Otherwise, clear it.
Taxonomy source: Taxonomy rules have been pre-defined for many popular databases. If you are adding a new database,
and appropriate taxonomy rules are not defined, refer to Chapter 9 of the Mascot Setup and Administration manual for further
information.
If you don't want to use taxonomy, select '--- None ---'. This will speed up the time taken to bring the database on-line.
Parse Rules: Apart from starting with a 'greater than' symbol, the precise syntax of the FASTA title line
varies from database to database. For this reason, Mascot uses Basic Regular Expressions to define how the accession
string and the description text should be parsed from the FASTA title line.
Two or three fields are used to select the regular expressions used to parse information from the database files.
The first rule defines how to parse an accession string from the FASTA file title line. The second, how to parse a
description string from the FASTA file title line. The third, how to parse an accession string from a local reference
file. If there is no local reference file, the third rule should be set to '--- no local reference file ---'.
Basic regular expressions are described in Appendix A of the Mascot Setup and Administration manual.
Source and parse rules for full text report:
This section defines how Mascot can retrieve a full
text report (references, feature table, etc.) for use in 'Protein View'. If full text is not required or not
available, just leave all fields blank, and choose '--- no full text report ---' in the drop down list.
Host: The report can come from a local reference file, or from a remote resource such as the NCBI Entrez
server. In the case of a local file, specify localhost. Otherwise, specify the remote host (webserver) from which
a report can be obtained by HTTP.
Port: Always specify 80 unless a remote host (webserver) is known to operate on a non-standard port.
Path: For a remote host, this path completes the URL required to retrieve a report. For localhost,
specify the path to ms-getseq.exe followed by the arguments required to retrieve a report.
#ACCESSION# is a placeholder for an accession string. Folders must be delimited by forward slashes,
even on a Windows system.
Parse rule: This parse rule is used to select the text that will appear in 'Protein View'.
If the host is a remote host, you'll probably want to remove the HTML tags.
Testing the definition
When the database definition appears to be complete, press the 'Test this definition' button. This will
run a number of tests on the consistency and syntax of the various settings. The script will also read the
first five and last five entries from the FASTA file (and local reference file if defined) and test the
parse rules. The test report for SwissProt should look something like this:
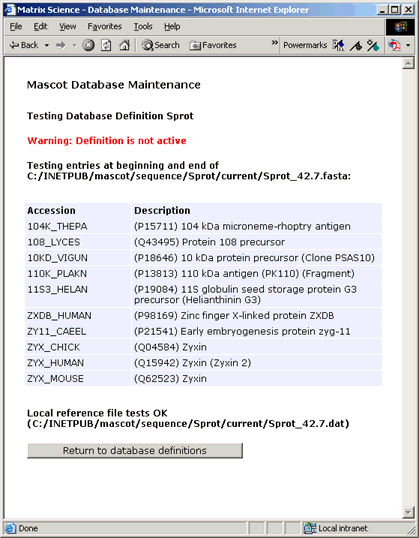
If a remote source was specified for the full text report, this will also be tested for the first entry in the database.
If any problems are encountered, there will either be explicit error messages, or the failure will be apparent from the
test results.
Saving the changes
Use the button at the bottom of the test report to return to the database definitions form and choose 'Apply'.
All being well, you can then follow the link to the Mascot database status page and monitor progress as the new database
is brought on-line.
|