Sequence Database Setup: Generic Database
Overview
A very simple configuration is sufficient for the common case of a sequence database where all the entries have the same taxonomy and
there is no full text reference file. To add such a database to Mascot, the requirements are just:
- You need a local copy of the database in Fasta format
- Each sequence must have a unique identifier / accession string
Download
A local copy of the Fasta format database is required. Often, this will be sequences representing the genome of a single organism,
and there will be a choice of files. For example, TIGR
has a list of completed microbial genomes. Follow the
links for Helicobacter pylori, and the following files are available for download:
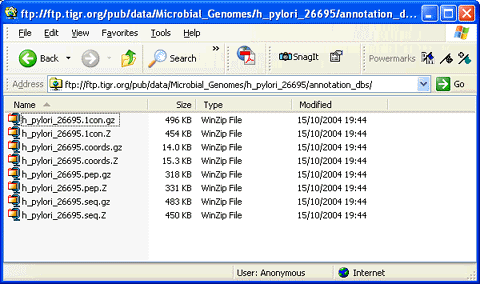
(Note that there are really only 4 different files, but each one has been compressed using two different compression tools).
This is pretty typical, and the files correspond to:
- Assembled genomic DNA sequence - h_pylori_26695.1con
- Nucleic acid coding sequences - h_pylori_26695.seq
- Coding sequences translated to proteins - h_pylori_26695.pep
- Table of co-ordinates for the coding sequences in the assembled chromosome - h_pylori_26695.coords
If you are confident that the coding sequences and reading frames have been
identified correctly, then the pep file would be the first choice for Mascot. It is possible to use the seq file, but this will
result in slower searches, because Mascot has to translate each sequence in all six reading frames.
If you are not confident that the coding sequences and reading frames have been identified correctly, then you might
wish to search the genomic DNA directly. Helicobacter pylori has a single chromosome, so h_pylori_26695.1con
contains just one sequence, of length 1,667,867 bases. This is not ideal for a Mascot search, because it would make the reports
too unwieldy. For efficient searching, genomic DNA needs to be chopped into shorter sequences, with small overlaps
to ensure no peptides are lost because they span a boundary. This is not a completely trivial task if
you want to maintain the original forward and reverse frame numbering from chunk to chunk. A simple perl utility to split
a long sequence can be downloaded here. Usage information can be obtained by
executing it with no arguments.
The relative merits of searching protein, EST and DNA sequences are discussed in TRENDS in Biotechnology 19(10)
S17-S22(2001) pdf.
Parse Rules
To decide on a suitable parse rule, you need to examine the title lines. If the database file is a large one, it may not be a
good idea to open it in a standard word processor or text editor. Most platforms support a command line utility called more
that can be used to browse a file of any size. In the case of h_pylori_26695.pep, the first few lines look like this:
>HP0001 hypothetical protein {Helicobacter pylori 26695}
MATRTQARGAVVELLYAFESGNEEIKKIASSMLEEKKIKNNQLAFALSLFNGVLEKINEI
DALIEPHLKDWDFKRLGSMEKAILRLGAYEIGFTPTQNPIIINECIELGKLYAEPNTPKF
LNAILDSLSKKLTQKPLN
>HP0002 riboflavin synthase beta chain (ribE) {Helicobacter pylori 26695}
MQIIEGKLQLQGNERVAILTSRFNHIITDRLQEGAMDCFKRHGGDEDLLDIVLVPGAYEL
PFILDKLLESEKYDGVCVLGAIIRGGTPHFDYVSAEATKGIAHAMLKYSMPVSFGVLTTD
NIEQAIERAGSKAGNKGFEAMSTLIELLSLCQTLKG
>HP0003 3-deoxy-d-manno-octulosonic acid 8-phosphate synthetase (kdsA) {Helicobacter pylori 26695}
MKTSKTKTPKSVLIAGPCVIESLENLRSIATKLQPLANNERLDFYFKASFDKANRTSLES
YRGPGLEKGLEMLQTIKEEFGYKILTDVHESYQASVAAKVADILQIPAFLCRQTDLIVEV
A simple rule that takes everything between the ">" symbol and the first space as the accession will work.
Everything after the first space can be treated as the description. These rules are pre-defined in mascot.dat as rules 4 and 5.
">\([^ ]*\)"
">[^ ]* \(.*\)"
Tips:
- If this rule looks like it should work, and doesn't, it may be because the space is actually a tab. If
this is the case, then you can use a character class that includes or excludes all the printing characters
">\([!-~]*\)"
">[!-~]*[^!-~]\(.*\)"
- Don't make a parse rule more precise than it needs to be. Looking at the first few lines, you might think that a
good rule would be:
">\(HP[0-9]*\)"
However, if you scroll down further, you'll find this title line:
>HP1203.5 preprotein translocase
The more picky rule will extract HP1203, giving a duplicate accession error message.
- (Mascot 2.0 and earlier only) If the longest accession string is more than 15 characters, you'll need to increase the default limit on the
accession string length. This is defined in mascot.dat by the parameter MaxAccessionLen. Set this to be a little longer
than the longest accession string.
Note: After changing MaxAccessionLen, you must re-compress all the active sequence databases.
To do this, set all databases to be inactive, then stop the Mascot service (Windows), or kill ms-monitor.exe (Unix). Delete the *.stats
file in the current directory of each sequence database, then re-start the Mascot service. Set the databases active
one at a time, waiting for each one to reach the "In use" state before moving on to the next.
- Several parse rules are pre-defined in mascot.dat. Experiment with these before writing a new one. If you have to
write a new one, remember that these are Basic Regular Expressions, as used in grep, not Extended Regular Expressions,
as used in Perl.
- If you need to edit a large sequence database file under Windows, you will need an editor that can edit the file without reading
it all into memory. One such editor is UltraEdit.
Configuration
For this example, h_pylori_26695.pep.gz was downloaded to a folder named
C:\Inetpub\MASCOT\sequence\h_pylori\current.
The file was decompressed using gzip and renamed to h_pylori.fasta. (Under Windows,
you can also use WinZip to decompress .Z and .gz files).
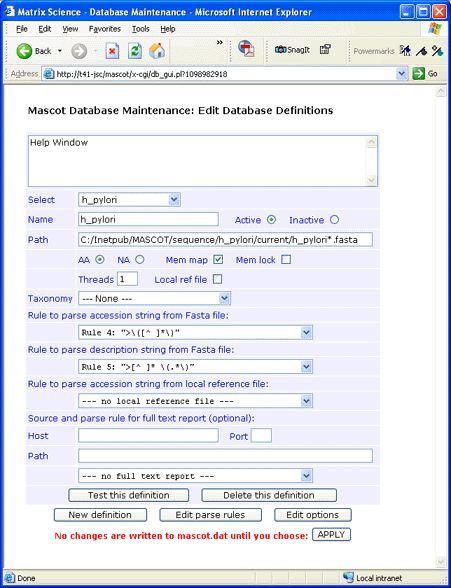
Tips:
- If you are setting up one of the nucleic acid files, the only change is to choose NA rather than AA
- In general, databases should be memory mapped but not memory locked
- The wild card in the database path is required
- The value of threads will default to the number of CPU's in your Mascot licence
- Always test a new definition before applying the changes to mascot.dat
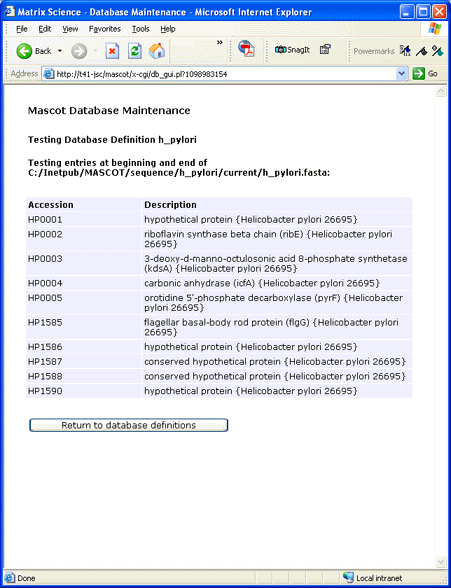
Assuming that there are no error messages from testing the database, choose Apply to save the new
configuration in mascot.dat. Then, follow the link to Database Status and verify that there are no
errors when the new database is compressed and tested. Once the database status reads "In Use",
the database is available for searching.
|