Sequence Database Setup: General Procedure
1. Choose a name for the Database
Give the database a short, descriptive name. This is the name that will appear in the
drop-down list in the search form, so you don't want to write an essay. Note that database
names are case sensitive.
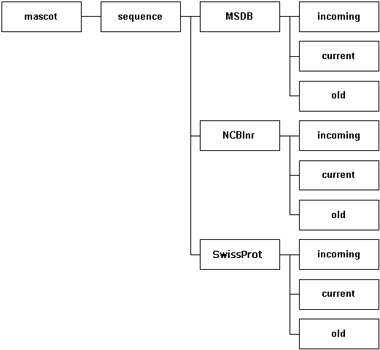
2. Create a local directory structure
The recommended arrangement is to have a dedicated directory for each database. Within this directory
are three sub-directories. The incoming directory provides a workspace for downloading and processing a new database file.
The current directory contains the active database, and this is where Mascot Monitor creates
the compressed files that will be memory mapped. The old directory is where the immediate past database files
are archived … just in case.
Points to watch:
- Giving the database and the directory the same name is usually a good idea, but is not
a requirement.
- There is no requirement for all the database directories to be placed in the mascot/sequence
directory.
- Under Windows, path and file names are not case sensitive, but it is safer to treat them as
if they were.
- Under Unix, links provide great flexibility, and the files or directories for a given database
can be located wherever convenient. If the Fasta file is actually a link, then Mascot will create the compressed
files in the directory containing the link, not in the target directory containing the Fasta file.
If you want the compressed files to be on a remote drive, you can do this by making a link at the
directory level. However, ensure that the network bandwidth is sufficient, and that the operating system
supports memory mapping of NFS mounted files.
- Mascot does not support Windows UNC paths.
3. Download the database files
Download at least one release of the database manually, so as to verify the filenames and URLs. If the
database is not pre-defined in the Mascot help text or in the database update script, make
careful notes of which files are required, where they come from, and any processing that is required.
Mascot can search any FASTA format sequence database. The FASTA format is extremely simple.
Each entry consists of a one line title followed by one or more lines containing the
sequence data in 1 letter code. FASTA databases can contain either amino acid sequences or nucleic
acid sequences, but not both. Nucleic acid databases are translated on the fly by Mascot in all six
reading frames.
The FASTA title line begins with a "greater than" character, followed by one or more accession strings,
and an optional text string describing the entry. Apart from the use of the "greater than" character,
the precise syntax of the title line is not defined. The title line is delimited from
the sequence that follows by a platform dependent new line character.
Line lengths vary between databases;
anything from 60 characters to a thousand or more. Mascot can handle lines up to 50,000 characters long.
The end of a sequence is indicated when the following line is either a new title line or the end of the file.
Some databases come with a "reference" file, containing annotation text and cross-reference information
in addition to the sequence. An example would be the
SwissProt Dat file.
Mascot may be able to use the reference file to get more accurate taxonomy information. It can
also incorporate the full text for an entry in the Protein View report. Even when a full text file is not available for download,
Mascot may be able to retrieve equivalent text from a remote HTTP server, such as NCBI's
Entrez or an
SRS server.
If database entries contain taxonomy information, Mascot can use this as a filter during a search. Many of the
most popular databases, such as SwissProt and NCBI nr, include taxonomy. To determine taxonomy accurately,
Mascot requires database specific supporting files. Details of these can be found in the help pages
for the individual databases. Note that these supporting files have to be downloaded into the taxonomy
directory, not into the sequence database directory. Also, some files need to be unpacked (using tar) as well as
uncompressed.
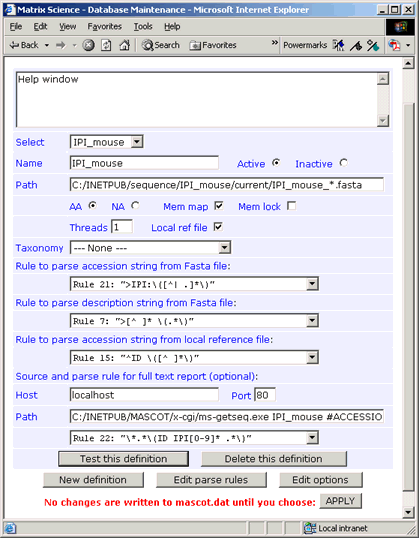
4. Configure the database
The easiest way to add a new database to Mascot is by using the Database
Maintenance script, accessible from a hyperlink on the Mascot home page, (Intranet only).
5. Bring the Database on-line
Once you 'Apply' a new definition, Mascot Monitor will look to see if there is a Fasta file that matches the
specified path. If so, it will begin to compress the Fasta file, so as to minimise the memory requirements.
If taxonomy has been defined for the database, Monitor will also create a taxonomy index.
Once this is complete, the new database is tested by running a standard search. If this succeeds, the new database
becomes available for general use.
6. Updates
Most sequence databases are growing rapidly, and you'll want to update the files regularly. Whenever Mascot Monitor sees a new
Fasta file that matches the wildcard path, it will automatically swap to the new database, as described below.
However, downloading and processing database updates by hand is tedious. Once the general procedure has been verified, it can
be automated using the Database Update script.
Databases can then be updated as often as you wish, with no disruption for Mascot users. Whenever
Monitor sees a new Fasta file that matches the defined path, the new database is compressed and tested.
If errors are detected in the new database, the database exchange process is abandoned.
Assuming the test
is successful, all new searches are performed against the new database, while searches that are in
progress against the old database are allowed to continue. Once the final search against the old database
is complete, it is unmapped from memory and the files moved to the "old" directory. The
new database is then memory mapped and the system becomes ready for the next update cycle.
|