Quantitation: emPAI protocol
The Exponentially Modified Protein Abundance
Index (emPAI) offers approximate, label-free, relative quantitation of the
proteins in a mixture based on protein coverage by the peptide matches in a database
search result. Developed by Ishihama and colleagues, the key publication is
Ishihama,
Y., et al., Exponentially modified protein abundance index (emPAI)
for estimation of absolute protein amount in proteomics by the number of
sequenced peptides per protein, Molecular & Cellular Proteomics 4 1265-1272 (2005)
Unlike the other quantitation protocols, the information required for emPAI is always
present in a search result, and there are no parameter settings, so
emPAI is "always on", as long as the MS/MS search contains at least 100 spectra.
The formula is very simple: 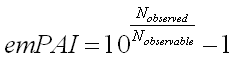
Where Nobserved is the number of experimentally observed
peptides and Nobservable is the calculated number of observable peptides for each protein.
The tricky bit is deciding
what to include and what to exclude in these two counts.
The number of observed peptides
The count of observed peptides only includes peptide matches with scores at or above the homology threshold,
or the identity threshold, if there is no homology threshold. Ishihama et. al. obtained best proportionality
for a standard protein mixture by counting unique parent ions,
including different charge states from the same peptide sequence.
We have followed this same rule.
The number of observable peptides
To estimate the number of observable peptides, Ishihama et. al. performed explicit
in silico digests of the protein sequences. The peptide list
was then filtered to exclude peptides outside the mass spectrometer
scan range and the observed nano-LC retention time range.
For reasons of speed, we prefer to make a calculated estimate of
the number of observable peptides based on the protein mass, the average amino acid composition of
the database, and the enzyme specificity. The error of doing this is negligible
compared with other sources of uncertainty:
- It isn't practical to filter by retention time, because this information is usually unavailable
- The mass range of the instrument has to be estimated from the range of precursors found in the data set
- Mass range filtering is by Mr, rather than m/z
- The digest is assumed to be a limit digest
- No obvious way to extend the calculation to semi-specific or non-specific digests
In the
supplementary
material for Ishihama et. al., there is a worked example for
human serum albumin which resulted in a count of 34 for the observable peptides
in the Mr range 700 to 2800 and the retention time range 40 to 150 minutes.
The enzyme was strict trypsin and no missed cleavages were allowed. The number of
peptides estimated by the routine used here, (&emPAI_digest), is 35.
Click here for an example of emPAI. We are grateful to Dr Jyoti Choudhary of the
Sanger Institute
for this small, LC-MS/MS data set from a human cell lysate
acquired using a Waters QTof. Notice how hits 14 and 15 both have the same number of
matches, but KPYM_HUMAN has the smaller emPAI value because Nobservable will be some 3 times
larger than for BASI_HUMAN.
|